|
Interaction Types
How to use PICI server?
How to interpret the data file?
Release notes
Hydrogen Bond
Default parameters for PICI server
|

D-H···Π Hydrogen bond |
| D-H···A |
dD-A (Å) |
∠D-H-A |
| N-H···O |
≤ 3.5 (1) |
≥ 110 ° (2) |
| N-H···N |
≤ 3.5 (1) |
| N-H···S |
≤ 4.1 (3) |
| O-H···N |
≤ 3.5 (1) |
| O-H···O |
≤ 3.5 (1) |
| O-H···S |
≤ 4.1 (3) |
| S-H···O |
≤ 4.3 (3) |
| S-H···N |
≤ 4.3 (3) |
| S-H···S |
≤ 4.5 (3) |
| C-H···O |
≤ 3.5 (4) |
| C-H···N |
≤ 3.5 (4) |
| C-H···S |
≤ 4.1 (3) |
| N-H···π |
< 4.3 (5) |
| O-H···π |
< 4.3 (5) |
| S-H···π |
≤ 4.5 (3) |
| C-H···π |
≤ 4.5 (6-8) |
Reference:
1. Baker, E.N., Hubbard, R.E. (1984) Hydrogen bonding in globular proteins. Prog Biophys Mol Biol, 44, 97-179.
2. Arunan, E., Desiraju, G.R., Klein, R.A., et al. (2011) Definition of the hydrogen bond (IUPAC Recommendations 2011). Pure and Applied Chemistry, 83, 1637-1641.
3. Zhou, P., Tian, F., Lv, F., Shang, Z. (2009) Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins, 76, 151-63.
4. Weiss, M.S., Brandl, M., Suhnel, J., Pal, D., Hilgenfeld, R. (2001) More hydrogen bonds for the (structural) biologist. Trends Biochem Sci, 26, 521-3.
5. Steiner, T., Koellner, G. (2001) Hydrogen bonds with pi-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J Mol Biol, 305, 535-57.
6. Brandl, M., Weiss, M.S., Jabs, A., Suhnel, J., Hilgenfeld, R. (2001) C-H...pi-interactions in proteins. J Mol Biol, 307, 357-77.
7. Steiner, T., Desiraju, G.R. (1998) Distinction between the weak hydrogen bond and the van der Waals interaction. Chemical Communications, 891-892.
8. Nishio, M., Umezawa, Y., Fantini, J., Weiss, M.S., Chakrabarti, P. (2014) CH-pi hydrogen bonds in biological macromolecules. Phys Chem Chem Phys, 16, 12648-83.
Aromatic-aromatic (Π-Π) interaction

Cation-aromatic interaction

Anion-aromatic interaction

Lone pair-aromatic (Lp-Π) interaction
How to use PICI server?
User can upload his/her protein structure file in pdb format at index page for calculation by PICI server. Pdb files can be uploaded to server by browsing to your file.
-
Click Browse / Choose File button (depends upon web browser type)
-
Click Submit button. (It will open the data processing page)
-
Select the radio button for your chain of interest.
-
Click Go button.
-
Calculation process will take few seconds for different types of interactions.
-
Result page will open after finishing the calculation. (description and image below.)
a. Buttons for different interactions.
b. Chain information of protein structure
c. Interacting residues in protein structure by JSmol visualizaton tool.
d. Interaction details in text.
e. Interacting residues in protein sequence.
|
 |
-
Click download button to download the interaction file in txt format.
How to interpret data file?
Interaction data files are being generated for each calculation and stored on the server.
(Note: Interaction files will be stored upto 15 days)
Users can download their data files by following URL: http://www.pici.bicpu.edu.in/download.php?accid=[ accession id ]&chain=[ chain id ]
Interaction File Description
Record: HBOND
Contains: Hydrogen bond description
| COLUMNS |
CONTENT |
| 0 - 6 |
bond type |
| 7 - 8 |
selected chain |
| 9 - 12 |
residue |
| 13 - 18 |
residue number |
| 19 - 20 |
donor/acceptor type |
| 21 - 26 |
atom |
| 27 - 28 |
other chain |
| 29 - 32 |
residue |
| 33 - 38 |
residue number |
| 39 - 40 |
donor/acceptor type |
| 41 - 46 |
atom |
| 47 - 52 |
donor-acceptor distance
(Å) |
| 53 - 58 |
hydrogen-acceptor distance(Å) |
| 59 - 66 |
d-h⋯a angle Θ° |
| 67 - 69 |
inter-chain type |
| 70 - 74 |
h-bond type |
| |
MM - Main chain to Main chain; SM - Main chain to Side chain; SS - Side chain to Side chain |
Example:
1 2 3 4 5 6 7 8
012345678901234567890123456789012345678901234567890123456789012345678901234567890
HBOND P SER 2 d OG A TYR 9 a O 3.35 2.63 145.30 SM OH⋯O
HBOND P SER 2 d N A TYR 9 a O 2.97 2.13 165.62 MM NH⋯O
HBOND P GLN 3 d N B GLU 74 a OE2 3.21 2.43 151.09 SM NH⋯O
HBOND P TYR 4 d N A ASN 62 a OD1 2.70 1.92 151.20 SM NH⋯O
HBOND P ARG 5 d N B TYR 30 a OH 2.99 2.21 151.97 SM NH⋯O
HBOND P SER 7 d N A ASN 69 a OD1 3.20 2.35 170.23 SM NH⋯O
Record: HPHOB
Contains: Hydrophobic interaction description
| COLUMNS |
CONTENT |
| 0 - 6 |
bond type |
| 7 - 8 |
selected chain |
| 9 - 12 |
residue |
| 13 - 18 |
residue number |
| 19 - 20 |
other chain |
| 21 - 24 |
residue |
| 25 - 30 |
residue number |
Example:
1 2 3
0123456789012345678901234567890
HPHOB A CYS 293 B TRP 539
HPHOB A LEU 295 B TRP 334
HPHOB A LEU 295 B TRP 539
HPHOB A PRO 297 B PRO 333
HPHOB A LEU 298 B TYR 434
HPHOB A LEU 298 B VAL 329
HPHOB A PHE 299 B LEU 331
HPHOB A PHE 299 B VAL 329
HPHOB A LEU 312 B TRP 539
Record: SSBOND / IONIC
Contains: Disulfide bond (SSBOND), Electrostatic interactions (IONIC)
| COLUMNS |
CONTENT |
| 0 - 6 |
bond type |
| 7 - 8 |
selected chain |
| 9 - 12 |
residue |
| 13 - 18 |
residue number |
| 19 - 23 |
atom |
| 24 - 30 |
atom number |
| 31 - 32 |
other chain |
| 33 - 36 |
residue |
| 37 - 42 |
residue number |
| 43 - 47 |
atom |
| 48 - 54 |
atom number |
| 55 - 60 |
distance (Å) |
Example:
1 2 3 4 5 6
0123456789012345678901234567890123456789012345678901234567890
SSBOND A CYS 293 SG 1024 B CYS 439 SG 2243 2.01
...
...
IONIC A ASP 225 OD2 479 B ARG 409 NH2 1986 2.73
IONIC A ASP 239 OD1 585 B LYS 413 NZ 2029 3.87
IONIC A GLU 241 OE2 606 B LYS 413 NZ 2029 3.10
IONIC A ASP 292 OD2 1018 B HIS 436 NE2 2223 3.13
IONIC A GLU 311 OE2 1176 B LYS 455 NZ 2365 2.83
IONIC A ASP 306 OD1 1132 B ARG 457 NH1 2379 3.93
Record: PIPI
Contains: Aromatic ring - aromatic ring interaction
| COLUMNS |
CONTENT |
| 0 - 6 |
bond type |
| 7 - 8 |
selected chain |
| 9 - 12 |
residue |
| 13 - 18 |
residue number |
| 19 - 20 |
other chain |
| 21 - 24 |
residue |
| 25 - 30 |
residue number |
| 31 - 36 |
distance (Å) [centroid - centroid] |
| 37 - 42 |
dihedral angle |
Example:
1 2 3 4
0123456789012345678901234567890123456789012
PIPI A TRP 4 B PHE 88 5.84 65.56
PIPI A TYR 316 B PHE 454 5.18 97.24
Record: CATPI / ANPI / SPI
Contains: Cation - aromatic ring (CATPI), Anion - aromatic ring (CATPI), Sulfur - aromatic ring (SPI) interactions
| COLUMNS |
CONTENT |
| 0 - 6 |
bond type |
| 7 - 8 |
selected chain |
| 9 - 12 |
residue |
| 13 - 18 |
residue number |
| 19 - 23 |
atom |
| 24 - 30 |
atom number |
| 31 - 32 |
other chain |
| 33 - 36 |
residue |
| 37 - 42 |
residue number |
| 43 - 47 |
atom |
| 48 - 54 |
atom number |
| 55 - 60 |
distance (Å) |
| 60 - 66 |
angle |
Example:
1 2 3 4 5 6
0123456789012345678901234567890123456789012345678901234567890123456
CATPI A ARG 296 CZ 1045 B TRP 539 RING - 4.74 64.56
CATPI A TRP 230 RING - B ARG 409 CZ 1984 3.49 24.49
CATPI A PHE 240 RING - B ARG 409 CZ 1984 4.02 33.68
...
...
ANPI A ASP 306 OD2 1133 B TRP 539 RING - 5.71 61.58
ANPI A GLU 309 OE1 1157 B TRP 539 RING - 5.98 77.39
ANPI A TYR 242 RING - B GLU 414 OE1 2037 4.66 69.21
ANPI A TYR 242 RING - B GLU 414 OE2 2038 5.59 84.88
ANPI A PHE 281 RING - B ASP 575 OD1 3319 5.78 89.89
...
...
SPI A PHE 277 RING - B CYS 439 SG 2243 5.26 68.49
SPI A TRP 285 RING - B MET 120 SD 1822 4.86 29.54
Release notes
PICI version 3.2 (released in Dec 2015)
-
Improved to calculate non-standard amino acids.
-
XML files are improved to handle data more efficiently.
-
Several bug fix for fast and accurate calculation.
-
Bug fix to remove false positive.
Older versions:
PICI version 3.1 (released in Sep 2015)
-
Calculation for interface area with NACCESS.
-
Improved interaction viewer with data display in tabular form or an interactive matrix.
-
Interaction files in downloadable XML and TXT file format.
-
Bug fix for hydrogen atom addition.
PICI version 2.9 (released in May 2015)
-
Hydrogen bond calculation now includes CH···O, CH···N, CH···S, OH···Π, NH···Π, SH···Π, CH···Π, types
-
Integration of HAAD and REDUCE programs to fix hydrogen atom positions in pdb files.
-
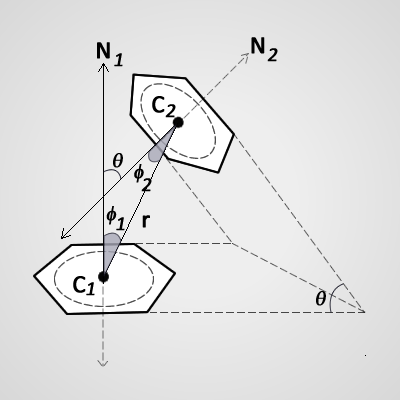
Calculation for aromatic ring – aromatic ring interactions with θ, Φ1 and Φ2 angles
-
Calculation for cation – aromatic ring interactions with θ
-
Calculation for anion – aromatic ring interactions with θ
-
Calculation for Lone pair – aromatic ring interactions with θ
-
Changed interaction file format
-
Changed JSmol view
-
Faster data processing and calculation
-
Visualization of interacting aminoacids on interaction matrix for each type of interactions
-
Improved user interface
PICI version 2.1 (released in 2014)
-
Hydrogen bond calculation now includes OH···O, OH···S, SH···O, SH···N, NH···S types apart from NH···O and OH···N types
-
For hydrophobic interactions hydrophobicities are based on solvent transfer free energies from octanol to water as mentioned by Brian W Mattews (1)
-
Calculation for aromatic ring – aromatic ring interactions
-
Calculation for cation – aromatic ring interactions
-
Calculation for anion – aromatic ring interactions
-
Calculation for sulfur – aromatic ring interactions
-
Changed interaction file format
-
Faster data processing and calculation
-
Integrated JSmol molecular visualization tool
-
Visualization of interacting aminoacids on an interaction matrix
-
Improved user interface
Reference:
1. Matthews, B.W. (2001) Hydrophobic Interactions in Proteins. Encyclopedia of life sciences. John Wiley & Sons, Inc.
PICI version 1.2 (released in 2011)
Integrated in PepBind database for calculating interaction interface in protein-peptide complexes.
-
NH···O and OH···N type hydrogen bond calculation.
-
For hydrophobic interactions hydrophobicities are based on Kyte-Doolittle hydropathy scale.
-
Calculation of disulfide bridges.
-
Calculation of ionic interactions (salt bridges).
-
Integrated Jmol molecular visualization tool.
-
Sequence display with highlighted interacting residues.
Citation: Das, A.A., Sharma, O.P., Kumar, M.S., Krishna, R., Mathur, P.P. (2013) PepBind: a comprehensive database and computational tool for analysis of protein-peptide interactions. Genomics Proteomics Bioinformatics, 11, 241-6.
|







